The intricate interplay between DNA and histone proteins results in the formation of chromatin, which performs a pivotal function in governing the expression of genes. The dynamic process of chromatin remodeling and modifications plays a decisive role in regulating the accessibility of DNA to the transcriptional machinery, thereby leaving an indelible imprint on gene expression. Thus, the isolation of chromatin assumes a paramount significance in unraveling the intricacies of gene expression regulation. The cutting-edge approach of Chromatin Isolation by RNA Purification (ChIRP) empowers researchers to capture the essence of specific chromatin regions, rendering it an indispensable technique for chromatin isolation. ChIRP achieves this feat through the employment of RNA probes that act as a beacon, leading researchers to the coveted target with unparalleled precision.
Chromatin Isolation by RNA Purification (ChIRP)
ChIRP is a pioneering method that hinges on the deployment of biotinylated RNA probes, which are designed to be complementary to the desired chromatin region. Once the RNA probe has successfully locked onto the target chromatin region, it is extracted and immobilized on streptavidin-coated magnetic beads. This intricate process confers a high degree of specificity and sensitivity to the approach, which can then be harnessed for downstream analyses. A range of analytical techniques, such as quantitative PCR (qPCR), Chromatin Immunoprecipitation-sequencing (ChIP-seq), and RNA sequencing (RNA-seq), can be utilized to study the captured chromatin region and deduce its functional significance.
The all-encompassing utility of ChIRP in illuminating the mysteries of gene expression regulation transcends species boundaries, as demonstrated by its successful application in a gamut of organisms, including yeast, mice, and humans. A compelling study centered on human cells employed ChIRP to isolate the promoter regions of diverse genes. The researchers noted a remarkable correlation between the histone acetylation levels and the gene's activity status, with the promoter regions of active genes showing augmented histone acetylation levels, while their inactive counterparts exhibited significantly reduced histone acetylation levels.

Main steps of ChIRP (Tian et al., 2020)
ChIRP-Seq: Combining ChIRP with Sequencing
The amalgamation of ChIRP with high-throughput sequencing has given rise to a prodigious tool known as ChIRP-Seq, which has revolutionized the field of chromatin analysis. ChIRP-Seq affords researchers the unparalleled ability to scrutinize chromatin regions bound by RNA probes in a genome-wide manner, obviating the need for targeted approaches. The initial step in ChIRP-Seq mirrors that of ChIRP, wherein RNA probes complementary to the target chromatin regions are deployed to capture the chromatin of interest. Once the chromatin has been apprehended, the RNA probes are systematically removed, paving the way for the next step - sequencing the DNA associated with the captured chromatin.
The resulting data derived from ChIRP-Seq is an extensive compendium of chromatin regions bound by RNA probes that can be analyzed with the aid of powerful bioinformatic tools. The information gleaned from ChIRP-Seq data can be leveraged to identify and catalogue the chromatin regions bound by the RNA probes with unprecedented accuracy and scale, empowering researchers to unravel the underlying mechanisms of gene expression regulation.
ChIRP-Seq Analysis: Identifying Chromatin Regions Bound by RNA Probes
The analysis of ChIRP-Seq data involves several steps, including alignment, peak calling, and downstream analysis. Alignment involves mapping the sequence reads to the reference genome. Peak calling involves identifying the chromatin regions that are significantly enriched in the ChIRP-Seq data. Downstream analysis involves interpreting the identified chromatin regions in the context of gene expression and regulation.
ChIRP-Seq data can provide insights into the regulatory mechanisms that control gene expression. In one study, researchers used ChIRP-Seq to study the role of long non-coding RNAs (lncRNAs) in the regulation of gene expression. They found that lncRNAs can interact with specific chromatin regions to regulate gene expression.
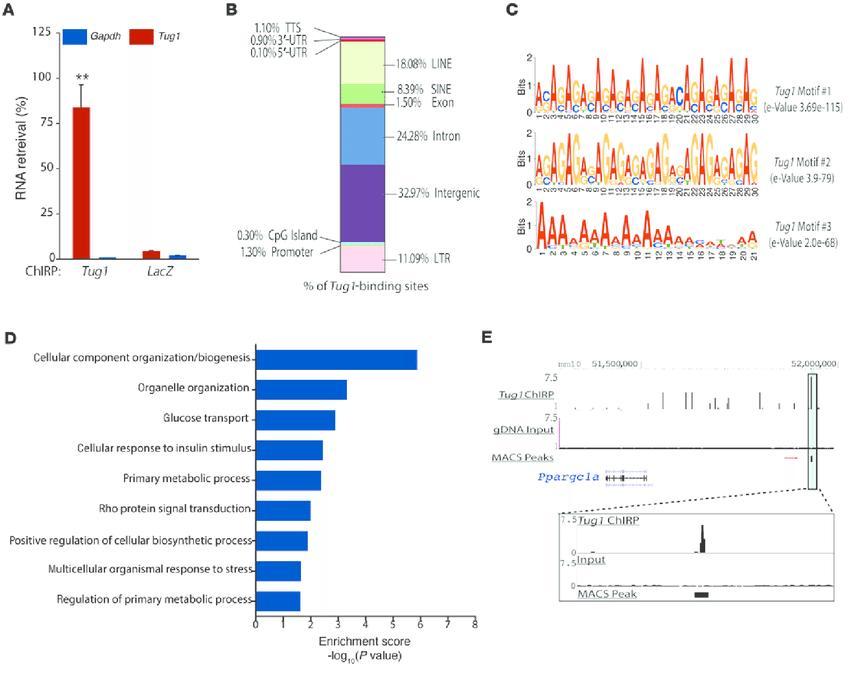
 ChIRP-seq analysis reveals genome-wide binding sites for Tug1, including a TBE upstream of the Ppargc1a promoter (Long et al., 2016)
ChIRP-seq analysis reveals genome-wide binding sites for Tug1, including a TBE upstream of the Ppargc1a promoter (Long et al., 2016)
References
- Tian, Changhai, and Guoku Hu. "Chromatin isolation by RNA purification (ChIRP) and its applications." Epigenetics Methods. Academic Press, 2020. 507-521.
- Long, Jianyin, et al. "Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy." The Journal of clinical investigation 126.11 (2016): 4205-4218.